全基因體關聯分析(GWAS)取樣策略
一、常見經濟作物樣本選擇
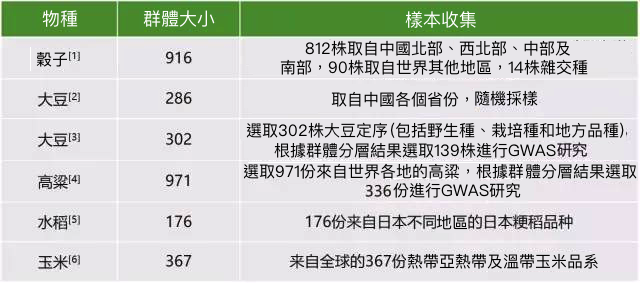
表1. 常見經濟作物樣本收集
二、常見哺乳動物物種選擇
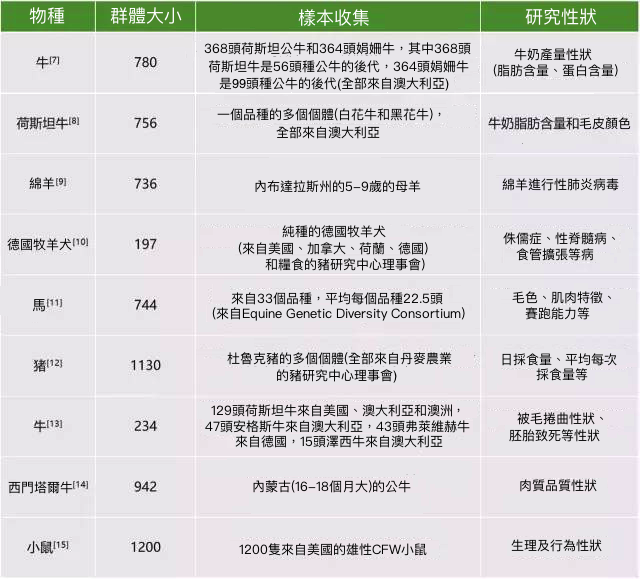
對於家禽而言,一般會選擇家系群體(全同胞家系或半同胞家系)。為了增加分析內容,可以構建多個家系群體進行研究。此外,盡量使群體所有個體生長環境以及營養程度保持一致家禽的年齡也盡量保持一致,這對錶型鑑定的準確性有很大的幫助。
表3.常見家禽類樣本收集

四、林木類樣本選擇
對於林木類,一般選擇同一物種的多個樣本,多個樣本做到表型豐富。
表4.林木類樣本收集

五、其他物種樣本選擇
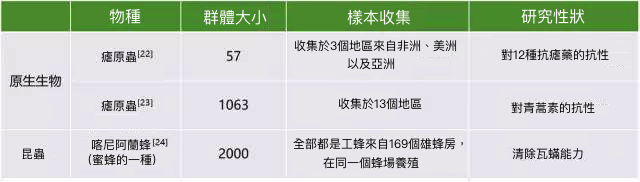
對於原生生物以及昆蟲等的取樣策略,可以參考表5中已發表的文獻。
表5.其他物種樣本收集
有這麼多個文獻支持,各位看官是不是已經明白了GWAS的如此取材呢?最後,小編再溫馨提示一句,根據文獻統計及項目經驗,一般來說,GWAS的樣本大小要不少於300個人是極好的。
參考文獻
[1] Jia G, Huang X, Zhi H, et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica)[J]. Nature Genetics, 2013, 45(8):957-61.
[2] Zhou L, Wang S B, Jian J, et al. Identification of domestication-related loci associated with flowering time and seed size in soybean with the RAD-seqgenotyping method[J]. Scientific reports, 2015, 5.
[3]Zhou Z, Jiang Y, Wang Z, et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean[J]. Nature Biotechnology, 2015, 33(4):408-414.
[4] MorrisG P, Ramu P, Deshpande S P, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum[J]. Proceedings of the National Academy of Sciences, 2013, 110(2): 453-458.
[5] Yano K, Yamamoto E, Aya K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice[J]. Nature Genetics, 2016, 48(8).
[6] Wang X, Wang H, Liu S, et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings[J]. Nature Genetics, 2016.
[7] Pryce J E, Bolormaa S, Chamberlain A J, et al. A validated genome-wide association study in 2 dairy cattle breeds for milk production and fertility traits using variable length haplotypes[J]. Journal of dairy science, 2010, 93(7):3331-3345.
[8] Hayes B J, Pryce J, Chamberlain A J, et al. Genetic architecture of complex traits and accuracy of genomic prediction:coat colour, milk-fat percentage, and type in Holstein cattle as contrastingmodel traits[J]. PLoS Genet, 2010, 6(9): e1001139.
[9] Heaton M P, Clawson M L, Chitko-Mckown C G,et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations[J].PLoS Genet, 2012, 8(1): e1002467.
[10] Tsai K L, Noorai R E, Starr-Moss A N, et al. Genome-wide association studies for multiple diseases of the German Shepherd Dog[J]. Mammalian Genome, 2012, 23(1-2): 203-211.
[11] Petersen J L, Mickelson J R, Rendahl A K, et al. Genome-wide analysis reveals selection for important traits in domestic horse breeds[J]. PLoS Genet, 2013,9(1): e1003211.
[12] Do D N, Strathe A B, Ostersen T, et al. Genome-wide association study reveals genetic architecture of eating behaviorin pigs and its implications for humans obesity by comparative mapping[J]. PLoS One, 2013, 8(8).
[13] Daetwyler H D, Capitan A, Pausch H, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic andcomplex traits in cattle[J]. Nature genetics, 2014, 46(8): 858-865.
[14] Wu Y, Fan H, Wang Y, et al. Genome-Wide Association Studies Using Haplotypes and Individual SNPs in Simmental Cattle[J]. PLoS One,2014,9(10): e109330.
[15] Parker C C, Gopalakrishnan S, Carbonetto P,et al. Genome-wide association study of behavioral, physiological and gene expression traits in outbred CFW mice[J]. Nature Genetics, 2016.
[16] Gu X, Feng C, Ma L, et al. Genome-wide association study of body weight in chicken F2 resource population[J]. PLoS One, 2011, 6(7): e21872.
[17] Xie L, Luo C, Zhang C, et al. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits[J]. PLoS One, 2012, 7(2): e30910.
[18] Liu R, Sun Y, Zhao G, et al. Genome-Wide Association Study Identifies Loci and Candidate Genes for Body Composition and Meat Quality Traits in Beijing-You Chickens[J]. Plos One, 2012, 8(4):-.
[19] Evans L M, Slavov G T, Rodgers-Melnick E, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations[J]. Nature genetics, 2014.
[20] Porth I, Klapšte J, Skyba O, et al. Genome‐wide association mapping for wood characteristics in Populus identifiesan array of candidate single nucleotide polymorphisms[J]. New Phytologist,2013, 200(3): 710-726.
[21] Van Tyne D, Park D J, Schaffner S F, et al. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum[J]. PLoS Genet, 2011, 7(4): e1001383.
[22] Ke C, Zhou Z, Qi W, et al. Genome-wide association study of 12 agronomic traits in peach[J]. Nature Communications,2016, 7:13246.
[23] Miotto O, Amato R, Ashley E A, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum[J]. Nature genetics, 2015, 47(3): 226-234.



